Why Write a File Ingest Module?
- Autopsy hides the fact that a file is coming from a file system, was carved, was from inside of a ZIP file, or was part of a local file. So, you don't need to spend time supporting all of the ways that your user may want to get data to you. You just need to worry about analyzing the content.
- Autopsy displays files automatically and can include them in reports if you use standard blackboard artifacts (described later). That means you don't need to worry about UIs and reports.
- Autopsy gives you access to results from other modules. So, you can build on top of their results instead of duplicating them.
Ingest Modules
For our first example, we're going to write an ingest module. Ingest modules in Autopsy run on the data sources that are added to a case. When you add a disk image (or local drive or logical folder) in Autopsy, you'll be presented with a list of modules to run (such as hash lookup and keyword search).
Those are all ingest modules. We're going to write one of those. There are two types of ingest modules that we can build:
- File Ingest Modules are the easiest to write. During their lifetime, they will get passed in each file in the data source. This includes files that are found via carving or inside of ZIP files (if those modules are also enabled).
- Data Source Ingest Modules require slightly more work because you have to query the database for the files of interest. If you only care about a small number of files, know their name, and know they won't be inside of ZIP files, then these are your best bet.
For this first tutorial, we're going to write a file ingest module. The second tutorial will focus on data source ingest modules. Regardless of the type of ingest module you are writing, you will need to work with two classes:
- The factory class provides Autopsy with module information such as display name and version. It also creates instances of ingest modules as needed.
- The ingest module class will do the actual analysis. One of these will be created per thread. For file ingest modules, Autopsy will typically create two or more of these at a time so that it can analyze files in parallel. If you keep things simple, and don't use static variables, then you don't have to think about anything multithreaded.
Getting Started
To write your first file ingest module, you'll need:
- An installed copy of Autopsy available from SleuthKit
- A text editor.
- A copy of the sample file ingest module from Github
Some other general notes are that you will be writing in Jython, which converts Python-looking code into Java. It has some limitations, including:
- You can't use Python 3 (you are limited to Python 2.7)
- You can't use libraries that use native code
But, Jython will give you access to all of the Java classes and services that Autopsy provides. So, if you want to stray from this example, then refer to the Developer docs on what classes and methods you have access to. The comments in the sample file will identify what type of object is being passed in along with a URL to its documentation.
Making Your Module Folder
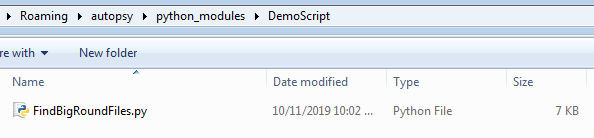
Every Python module in Autopsy gets its own folder. This reduces naming collisions between modules. To find out where you should put your Python module, launch Autopsy and choose the Tools -> Python Plugins menu item. That will open a folder in your AppData folder, such as "C:\Users\JDoe\AppData\Roaming\Autopsy\python_modules".
Make a folder inside of there to store your module. Call it "DemoScript". Copy the fileIngestModule.py sample file listed above into the this new folder and rename it to FindBigRoundFiles.py. Your folder should look like this:
Writing the Script
We are going to write a script that flags any file that is larger than 10MB and whose size is a multiple of 4096. We'll call these big and round files. This kind of technique could be useful for finding encrypted files. An additional check would be for entropy of the file, but we'll keep the example simple.
Open the FindBigRoundFiles.py file in your favorite python text editor. The sample Autopsy Python modules all have TODO entries in them to let you know what you should change. The below steps jump from one TODO to the next.
- Factory Class Name: The first thing to do is rename the sample class name from "SampleJythonFileIngestModuleFactory" to "FindBigRoundFilesIngestModuleFactory". In the sample module, there are several uses of this class name, so you should search and replace for these strings.
- Name and Description: The next TODO entries are for names and descriptions. These are shown to users. For this example, we'll name it "Big and Round File Finder". The description can be anything you want. Note that Autopsy requires that modules have unique names, so don't make it too generic.
- Ingest Module Class Name: The next thing to do is rename the ingest module class from "SampleJythonFileIngestModule" to "FindBigRoundFilesIngestModule". Our usual naming convention is that this class is the same as the factory class with "Factory" removed from the end.
- startUp() method: The startUp() method is where each module initializes. For our example, we don't need to do anything special in here. Typically though, this is where you want to do stuff that could fail because throwing an exception here causes the entire ingest to stop.
- process() method: This is where we do our analysis. The sample module is well documented with what it does. It ignores non-files, looks at the file name, and makes a blackboard artifact for ".txt" files. There are also a bunch of other things that it does to show examples for easy copy and pasting, but we don't need them in our module. We'll cover what goes into this method in the next section.
- shutdown() method: The shutDown() method either frees resources that were allocated or sends summary messages. For our module, it will do nothing.
The process() Method
The process() method is passed in a reference to an AbstractFile Object. With this, you have access to all of a file's contents and metadata. We want to flag files that are larger than 10MB and that are a multiple of 4096 bytes. The following code does that:
if ((file.getSize() > 10485760) and ((file.getSize() % 4096) == 0)):
Now that we have found the files, we want to do something with them. In our situation, we just want to alert the user to them. We do this by making an "Interesting Item" blackboard artifact. The Blackboard is where ingest modules can communicate with each other and with the Autopsy GUI. The blackboard has a set of artifacts on it and each artifact:
- Has a type
- Has a category
- Is associated with a file
- Has one or more attributes. Attributes are simply name and value pairs.
A list of standard artifact types can be found in the artifact catalog. It is important to note the catagory for the artifact you want to since this affects which method you will use to create the artifact.
For our example, we are going to make an artifact of type "TSK_INTERESTING_ITEM", which is an analysis result, whenever we find a big and round file. These are one of the most generic artifact types and are simply a way of alerting the user that a file is interesting for some reason. Once you make the artifact, it will be shown in the UI. The below code makes an artifact for the file and puts it into the set of "Big and Round Files". You can create whatever set names you want. The Autopsy GUI organizes Interesting Files by their set name.
art = file.newAnalysisResult(BlackboardArtifact.Type.TSK_INTERESTING_ITEM, Score.SCORE_LIKELY_NOTABLE,
None, "Big and Round Files", None,
Arrays.asList(
BlackboardAttribute(BlackboardAttribute.Type.TSK_SET_NAME,
FindBigRoundFilesIngestModuleFactory.moduleName,
"Big and Round Files"))).getAnalysisResult()The above code adds the artifact and a single attribute to the blackboard in the embedded database, but it does not notify other modules or the UI. Calling postArtifact() will let the tree viewer and other parts of the UI know that a refresh may be necessary, and passes the newly created artifacts to other modules that may do further processing on it.
blackboard.postArtifact(art, FindBigRoundFilesIngestModuleFactory.moduleName)
That's it. Your process() method should look something like this:
def process(self, file):
# Use blackboard class to index blackboard artifacts for keyword search
blackboard = Case.getCurrentCase().getSleuthkitCase().getBlackboard()
# Skip non-files
if ((file.getType() == TskData.TSK_DB_FILES_TYPE_ENUM.UNALLOC_BLOCKS) or
(file.getType() == TskData.TSK_DB_FILES_TYPE_ENUM.UNUSED_BLOCKS) or
(file.isFile() == False)):
return IngestModule.ProcessResult.OK
# Look for files bigger than 10MB that are a multiple of 4096
if ((file.getSize() > 10485760) and ((file.getSize() % 4096) == 0)):
# Make an artifact on the blackboard. TSK_INTERESTING_ITEM is a generic type of
# artifact. Refer to the developer docs for other examples.
art = file.newAnalysisResult(BlackboardArtifact.Type.TSK_INTERESTING_ITEM, Score.SCORE_LIKELY_NOTABLE,
None, "Big and Round Files", None,
Arrays.asList(
BlackboardAttribute(BlackboardAttribute.Type.TSK_SET_NAME,
FindBigRoundFilesIngestModuleFactory.moduleName,
"Big and Round Files"))).getAnalysisResult()
try:
# post the artifact for listeners of artifact events
blackboard.postArtifact(art, FindBigRoundFilesIngestModuleFactory.moduleName)
except Blackboard.BlackboardException as e:
self.log(Level.SEVERE, "Error indexing artifact " + art.getDisplayName())
return IngestModule.ProcessResult.OKSave this file and run the module on some of your data. If you have any big and round files, you should see an entry under the "Interesting Items" node in the tree.
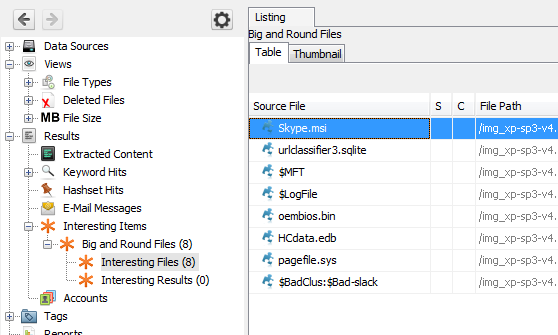
The full big and round file module along with test data can be found on github.
Debugging and Development Tips
Whenever you have syntax errors or other errors in your script, you will get some form of dialog from Autopsy when you try to run ingest modules. If that happens, fix the problem and run ingest modules again. You don't need to restart Autopsy each time!
The sample module has some log statements in there to help debug what is going on since we don't know of better ways to debug the scripts while running in Autopsy.